Note
Go to the end to download the full example code
Load OMF Project
Load and visualize an OMF project file
Originally from: https://opengeovis.github.io/omfvista/examples/load-project.html
# sphinx_gallery_thumbnail_number = 3
import omfvista
import pooch
import pyvista as pv
Load the project into an pyvista.MultiBlock dataset
url = "https://raw.githubusercontent.com/pyvista/vtk-data/master/Data/test_file.omf"
file_path = pooch.retrieve(url=url, known_hash=None)
project = omfvista.load_project(file_path)
print(project)
MultiBlock (0x7f2f38ffa040)
N Blocks 9
X Bounds 443941.105, 447059.611
Y Bounds 491941.536, 495059.859
Z Bounds 2330.000, 3555.942
Once the data is loaded as a pyvista.MultiBlock dataset from
omfvista, then that object can be directly used for interactive 3D
visualization from pyvista:
project.plot()
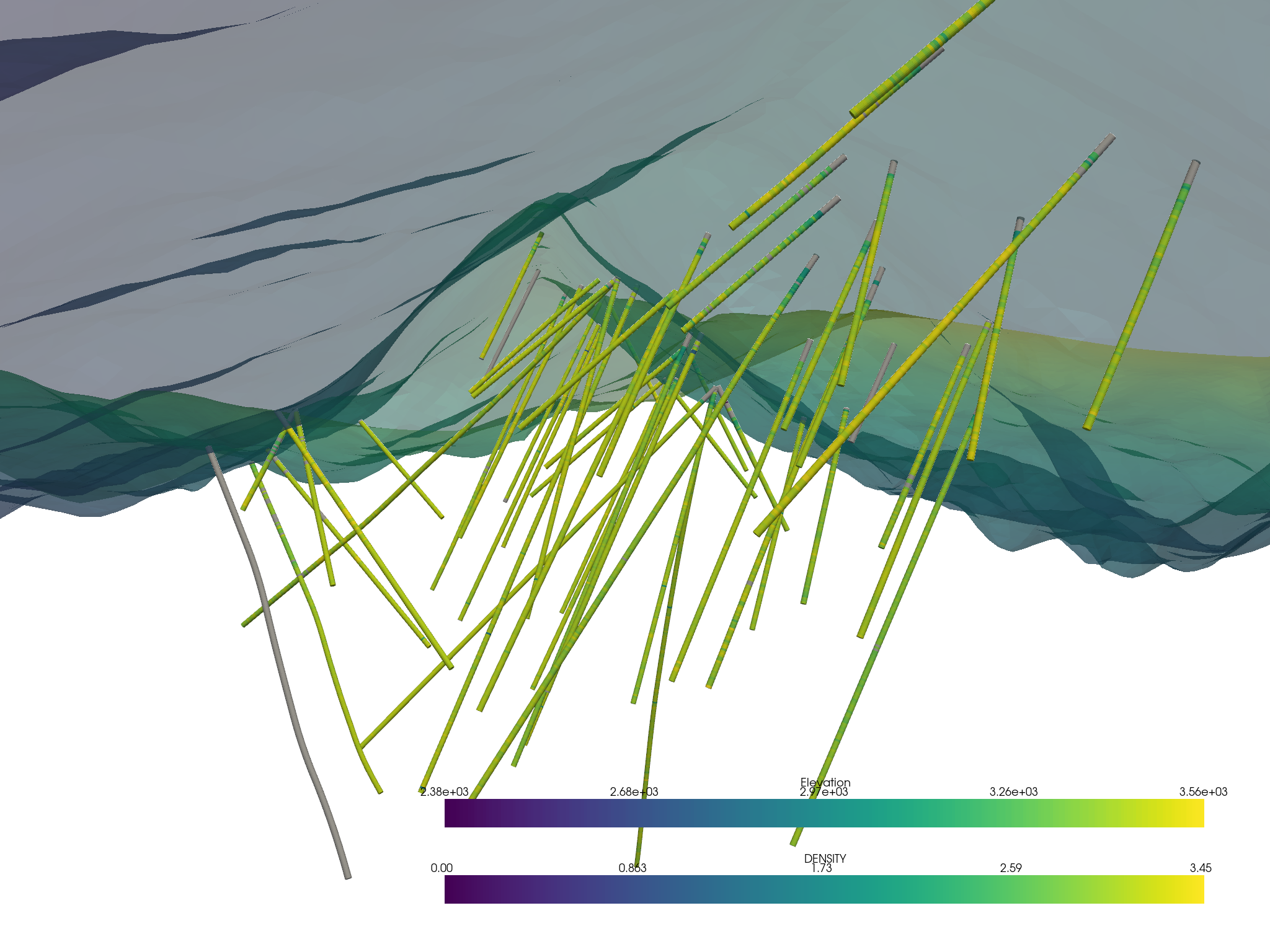
Or an interactive scene can be created and manipulated to create a compelling figure directly in a Jupyter notebook. First, grab the elements from the project:
# Grab a few elements of interest and plot em up!
vol = project["Block Model"]
assay = project["wolfpass_WP_assay"]
topo = project["Topography"]
dacite = project["Dacite"]
assay.set_active_scalars("DENSITY")
p = pv.Plotter()
p.add_mesh(assay.tube(radius=3))
p.add_mesh(topo, opacity=0.5)
p.camera_position = [
(445542.1943310096, 491993.83439313783, 2319.4833541935445),
(445279.0538059701, 493496.6896061105, 2751.562316285356),
(-0.03677380086746433, -0.2820672798388477, 0.9586895937758338),
]
p.show()
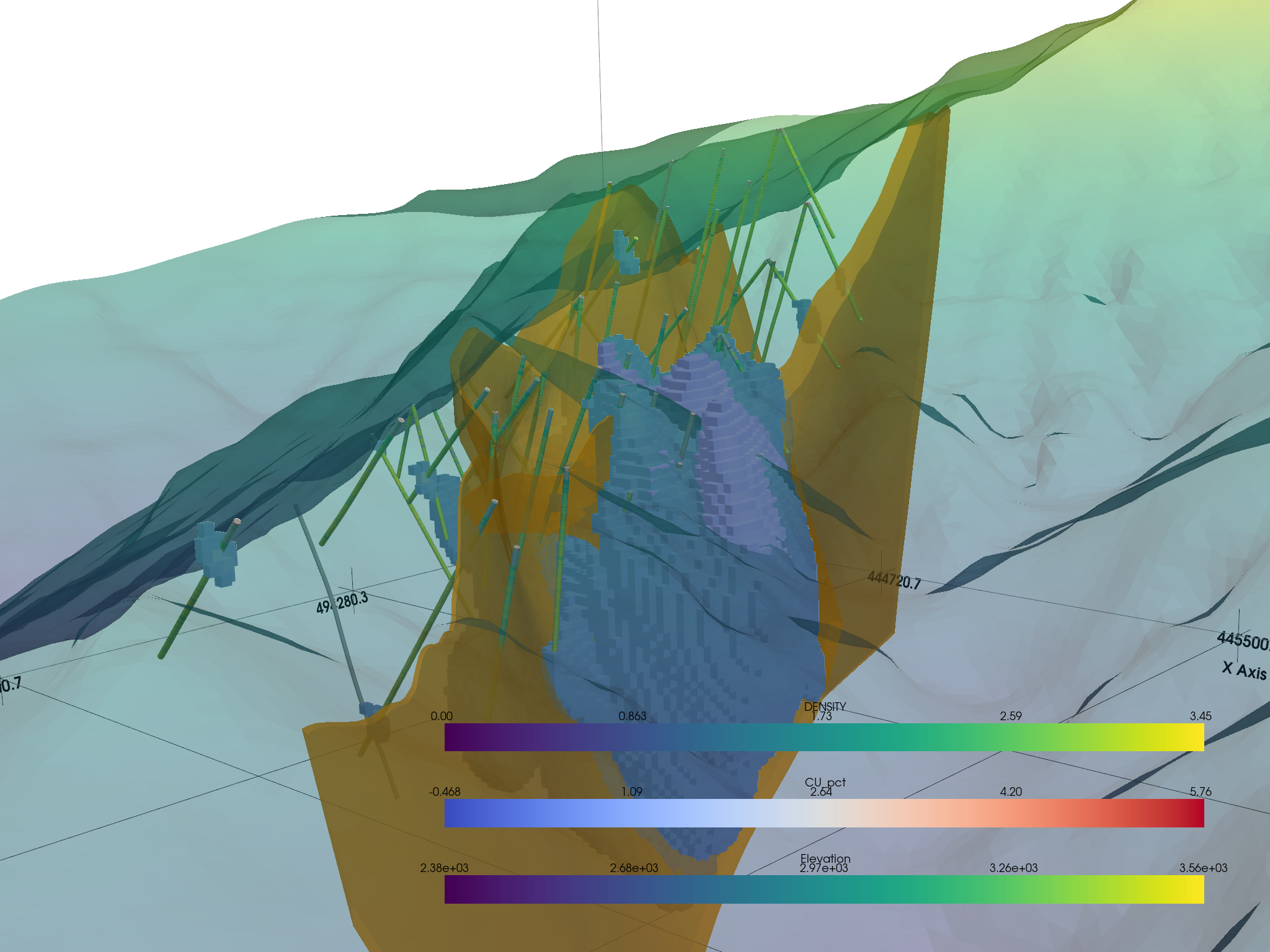
Then apply a filtering tool from pyvista to the volumetric data:
# Threshold the volumetric data
thresh_vol = vol.threshold([1.09, 4.20])
print(thresh_vol)
UnstructuredGrid (0x7f2f39bfa040)
N Cells: 92525
N Points: 107807
X Bounds: 4.447e+05, 4.457e+05
Y Bounds: 4.929e+05, 4.942e+05
Z Bounds: 2.330e+03, 3.110e+03
N Arrays: 1
Then you can put it all in one environment!
# Create a plotting window
p = pv.Plotter()
# Add the bounds axis
p.show_grid()
p.add_bounding_box()
# Add our datasets
p.add_mesh(topo, opacity=0.5)
p.add_mesh(
dacite,
color="orange",
opacity=0.6,
)
p.add_mesh(thresh_vol, cmap="coolwarm", clim=vol.get_data_range())
# Add the assay logs: use a tube filter that various the radius by an attribute
p.add_mesh(assay.tube(radius=3), cmap="viridis")
p.camera_position = [
(446842.54037898243, 492089.0563631193, 3229.5037597889404),
(445265.2503466077, 493747.3230470255, 2799.8853219866005),
(-0.10728419235836695, 0.1524885965210015, 0.9824649255831316),
]
p.show()
Total running time of the script: (0 minutes 17.137 seconds)